Offering a Suite of Clinical Grade Products for Any NGS Laboratory
Enable accurate, specialty testing for all patients.
Optimize treatment decisions and improve patient outcomes.
IVD Assays
Select Featured Research-Use-Only (RUO) Panels
The oncoReveal Solid Tumor v2 Panel is a robust 48-gene assay that simultaneously tests for key mutations present in solid tumors, including NSCLC, colorectal, melanoma, endometrial, pancreatic, GIST, bladder, thyroid, and gliomas. Additionally, genes with potential importance in immuno-oncology such as POLD1 and POLE are analyzed. The panel uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment.
oncoReveal Solid Tumor v2 Panel (48 genes)
The oncoReveal Multi-Cancer with CNV & RNA Fusion Panel is a combined DNA/RNA multi-cancer panel. The assay combines the DNA-based oncoReveal Multi-Cancer v4 with CNV Panel with the oncoReveal Multi-Cancer RNA Fusion v2 Panel allowing for joint sequencing. The assay uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment.
Variants and CNVs detected from DNA
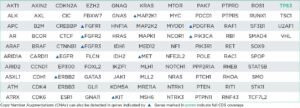
The oncoReveal Multi-Cancer v4 with CNV Panel is a robust NGS assay that interrogates genes of interest* across multiple solid tumor cancer types. The assay uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment. In addition to SNVs and indels, the assay detects CNVs in 14 genes.
oncoReveal Multi-Cancer v4 with CNV Panels (60 genes)
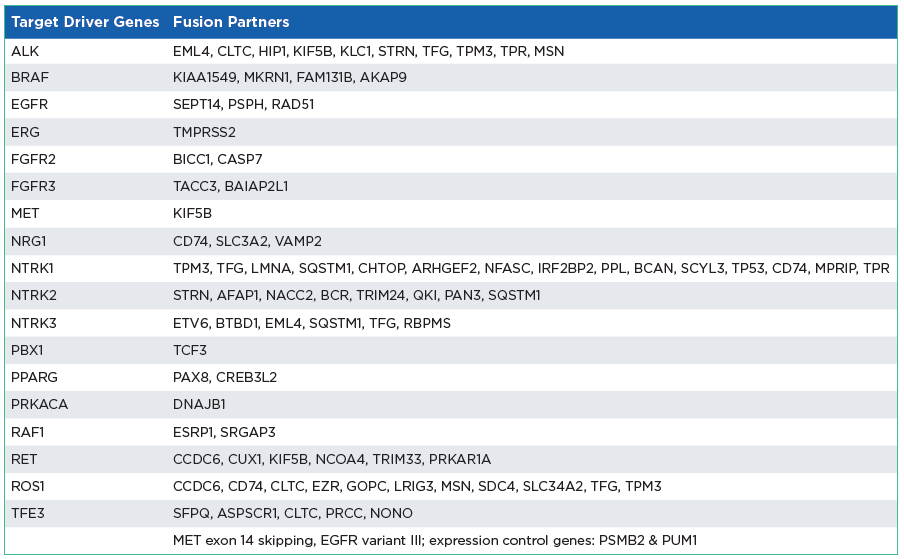
The oncoReveal Multi-Cancer RNA Fusion v2 Panel is a robust NGS assay that interrogates multiple gene rearrangement regions of interest* across multiple solid tumor cancer types. The assay uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment.
The oncoReveal Lung and Colon Cancer Panel is a robust NGS assay that interrogates 22 genes of interest* relevant to lung and colon cancer. The panel uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment.
oncoReveal Lung and Colon Cancer Panel (22 genes)
The oncoReveal Myeloid Panel is a robust NGS assay that interrogates 58 genes of interest* most relevant to myeloid cancers. The panel uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment.
oncoReveal Myeloid Panel (58 genes)
The oncoReveal Essential MPN Panel is a robust NGS assay that simultaneously tests for key mutations within the MPL, JAK2, and CALR genes. The panel uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment.
oncoReveal Essential MPN Panel
The oncoReveal BRCA1 & BRCA2 plus CNV Panel is designed for the detection of exon-level copy number variation (CNV) across the entire coding region of the BRCA1 and BRCA2 genes. The assay is designed with shorter amplicons to allow for detection of mutations from FFPE samples, while also being compatible with intact genomic DNA from blood samples. The assay sequences the full coding region plus 10bp of flanking introns. The panel utilizes proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment.
The oncoReveal Core LBx panel is a robust NGS assay that interrogates 104 genes of interest* across multiple solid tumor cancer types. The panel is specifically designed for cell-free DNA (cfDNA) extracted from plasma and can detect four types of variants: single nucleotide variants (SNVs), small insertion/deletion (indel) variants, copy number amplification (CNA), and microsatellite instability (MSI). The assay uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment.
oncoReveal Core LBx Panel (104 genes)
The oncoReveal Fusion LBx panel is a robust NGS assay that interrogates multiple gene rearrangement regions of interest across multiple solid tumor cancer types. The assay uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube enrichment of targets from cell-free RNA extracted from plasma.
oncoReveal Fusion LBx Panel
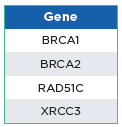
The oncoReveal 4 Gene Methylation Panel is a robust NGS assay that interrogates CpG sites within BRCA1, BRCA2, RAD51C, and XRCC3 that have undergone bisulfite conversion for methylome analysis. The assay uses proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based
library prep chemistry for efficient single-tube enrichment of targets.
oncoReveal 4 Gene Methylation Panel
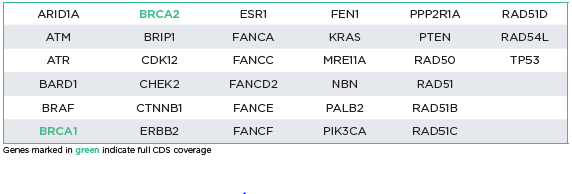
The oncoReveal HRD v2 Panel is designed for detection of SNVs, indels, and BRCA1 & BRCA2 exon level CNVs. 33 HRD-related genes can be assessed in either FFPE or blood samples. The panel utilizes proprietary Stem-Loop Inhibition-Mediated amplification (SLIMamp®) technology, a tiled amplicon-based library prep chemistry for efficient single-tube target enrichment.
9 Strathmore Rd., Natick MA, 01740